from_files#
- ethology.io.annotations.load_bboxes.from_files(file_paths, format, images_dirs=None)[source]#
Load an
ethologybounding box annotations dataset from a file.- Parameters:
file_paths (pathlib.Path | str | list[pathlib.Path | str]) – Path or list of paths to the input annotation files.
format ({"VIA", "COCO"}) – Format of the input annotation files.
images_dirs (pathlib.Path | str | list[pathlib.Path | str], optional) – Path or list of paths to the directories containing the images the annotations refer to. The paths are added to the dataset attributes.
- Returns:
A valid bounding box annotations dataset with dimensions image_id, space, id, and the following arrays:
position, with dimensions (image_id, space, id),
shape, with dimensions (image_id, space, id),
category, with dimensions (image_id, id) - optional,
image_shape, with dimensions (image_id, space) - optional.
The category array, if present, holds category IDs as 1-based integers, matching the category IDs in the input file. The dataset attributes include:
annotation_files: a list of paths to the input annotation files
annotation_format: the format of the input annotation files
map_category_to_str: a map from category ID to category name
map_image_id_to_filename: a map from image ID to image filename
images_directories: directory paths for the images (optional)
- Return type:
Notes
The image_id is assigned based on the alphabetically sorted list of unique image filenames across all input files. So if two images have the same filename but are in different input annotation files, they will be assigned the same image ID and their annotations will be merged.
The id dimension corresponds to the annotation ID per image. It ranges from 0 to the maximum number of annotations per image in the dataset. Note that the annotation IDs are not necessarily consistent across images. This means that the annotations with ID=3 in image t and image t+1 will likely not correspond to the same individual.
The space dimension holds the “x” and “y” coordinates.
Note that supercategories are not currently added to the xarray dataset, even if specified in the input file.
Examples
Load annotations from a single COCO file:
>>> from ethology.io.annotations import load_bboxes >>> ds = load_bboxes.from_files( ... file_paths="path/to/annotation_file.json", format="COCO" ... )
Load annotations from a single COCO file and specify the images directory:
>>> from ethology.io.annotations import load_bboxes >>> ds = load_bboxes.from_files( ... file_paths="path/to/annotation_file.json", ... format="COCO", ... images_dirs="path/to/images_dir", ... )
Load annotations from two VIA files and specify multiple image directories:
>>> from ethology.io.annotations import load_bboxes >>> ds = load_bboxes.from_files( ... file_paths=[ ... "path/to/annotation_file_1.json", ... "path/to/annotation_file_2.json", ... ], ... format="VIA", ... images_dirs=["path/to/images_dir_1", "path/to/images_dir_2"], ... )
Examples using from_files#
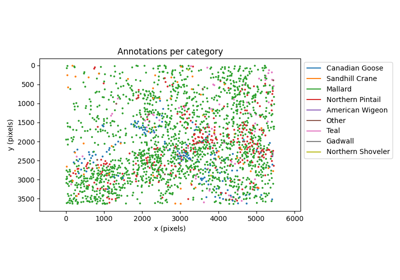
Inspect COCO annotations using ethology and movement